Alternative splicing events¶
Isotools defines alternative splicing events (ASE) with a graph based approach. The algorithm finds all ASE, classifies them by type and quantifies them with the PSI per sample. We distinguish 5 different event types: Exon Skipping(ES), Intron Retention (IR), Mutually Exclusive Exons(ME), 3’ Alternative Splicing (3AS), 5’ Alternative Splicing (5AS), Alternative First Exons (TSS), and Alternative Last Exons (PAS).
In this tutorial, we learn to use ASE to calculate 2D embeddings (PCA, UMAP), representing the relation of the samples with respect to alternative splicing. This representation may be a useful as a first step of the analysis, to validate hypothesis and check sample integrity.
To run this tutorial, download the transcriptome object file ‘PacBio_isotools_substantial_isotools.pkl’ from here to a subfolder ‘demonstration_dataset’.
[1]:
from isotools import Transcriptome
import matplotlib.pyplot as plt
path='demonstration_dataset'
isoseq=Transcriptome.load(f'{path}/PacBio_isotools_substantial_isotools.pkl')
[2]:
# compute the alternative splicng events
splice_events=isoseq.alternative_splicing_events()
# count the identified events by type
splice_events.splice_type.value_counts()
[2]:
TSS 391
PAS 254
ES 109
IR 78
5AS 54
3AS 37
ME 8
Name: splice_type, dtype: int64
PCA plots¶
[4]:
#plot PCA embedding
from isotools.plots import plot_embedding
plt.rcParams["figure.figsize"] = (10,10)
pca={}
f,axs=plt.subplots(4,2)
for ax,t in zip(axs.flatten(),['all','3AS','5AS','ES','IR','ME', 'TSS', 'PAS']):
pca[t]=plot_embedding(splice_events, ax=ax, labels=True, groups=isoseq.groups(), splice_types=t)
axs[0,0].legend(fontsize='medium', ncol=4,handleheight=2.4, labelspacing=0.05, bbox_to_anchor=(0, 1.1), loc='lower left')
plt.tight_layout()
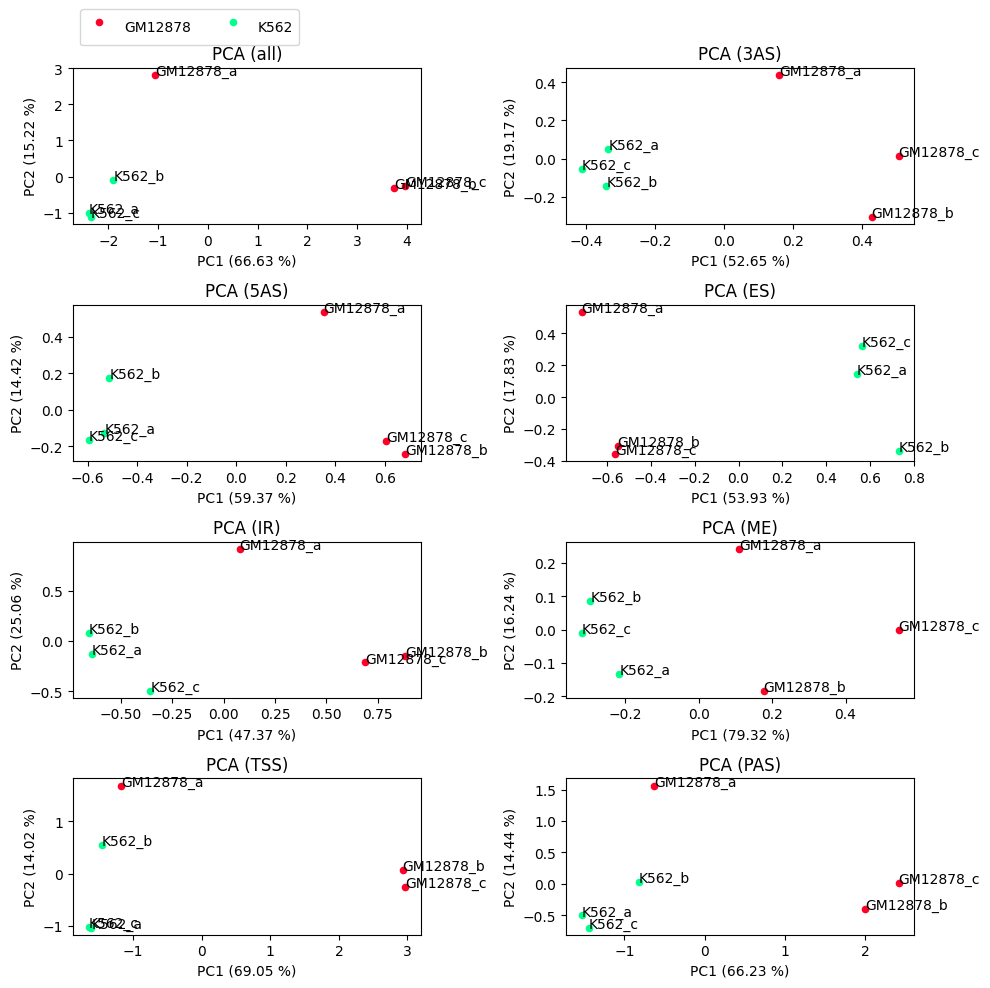
While the samples cluster as expected, e.g. the groups cluster together, it seems like sample GM12878_a is diverging from the two other samples of the same group.
[ ]: